The use of bioactive NPs dates back thousands of years and are prime candidates for the development of lead drugs. The ADD Hub has committed to identifying novel hit compounds that can be progressed to lead optimization in future projects. Through this research, we build on the extensive repertoire of SA & UK drug discovery expertise, with the aim of unlocking potential resources contained in natural marine and terrestrial biota.
Recent advances in state-of-the-art dereplication approaches, guided by machine learning & AI, and these advances have led to an increased focus in bacteria as producers of biological active metabolites. Entire, highly complex microbial communities exist in symbiosis with host sponges. The microbial communities of sponges are often specific to the host, and provide a multitude of services, such as protection from predators or invaders. Genes encoding secondary metabolites are often grouped together in a cluster and controlled by a single regulator. Orphan gene clusters are biosynthetic pathways for which the product has yet to be discovered as the molecular signal necessary for production is commonly absent in a laboratory setting. We are exploring the hidden potential of microbes by mining metagenomic datasets to identify interesting biosynthetic gene clusters (BGCs), which allows the assessment of culture-independent meta-omics to better understand the physiology and biochemical requirements of uncultivated bacteria.
The marine environment, which is largely unexplored, hosts a rich species diversity with unique adaptations, producing attractive and important secondary metabolites. Porifera & Cnideria phyla account for the majority (60 %) of marine bioactive compounds. Sponges are incapable of escaping predators or avoiding disease and fouling. Instead, they produce a variety of toxic secondary metabolites to ward off predators, compete with other sponges for living space, and to protect against infection. Among these secondary metabolites, we observe a wide diversity of structure and function that fall into clinically important chemical classes, including quinones, alkaloids, polyketides, terpenes, and peptides.
NPs are an unprecedented starting point for antibiotic discovery; NPs and their synthetic analogues are the source of >80% of all clinically-utilized antibiotics. NPs are privileged scaffolds, honed by evolution to possess many of the requisite properties of antibacterial drugs - properties often frustratingly lacking in synthetic compound libraries. Evolutionary conserved targets, which are less prone to mutation and the inadvertent development of antibiotic resistance, are becoming increasingly important targets in antimicrobial drug discovery. Interesting compounds are characterized by ADMET analysis, and analogues of interesting compounds are screened for improved antimicrobial activity and low eukaryote cytotoxicity.
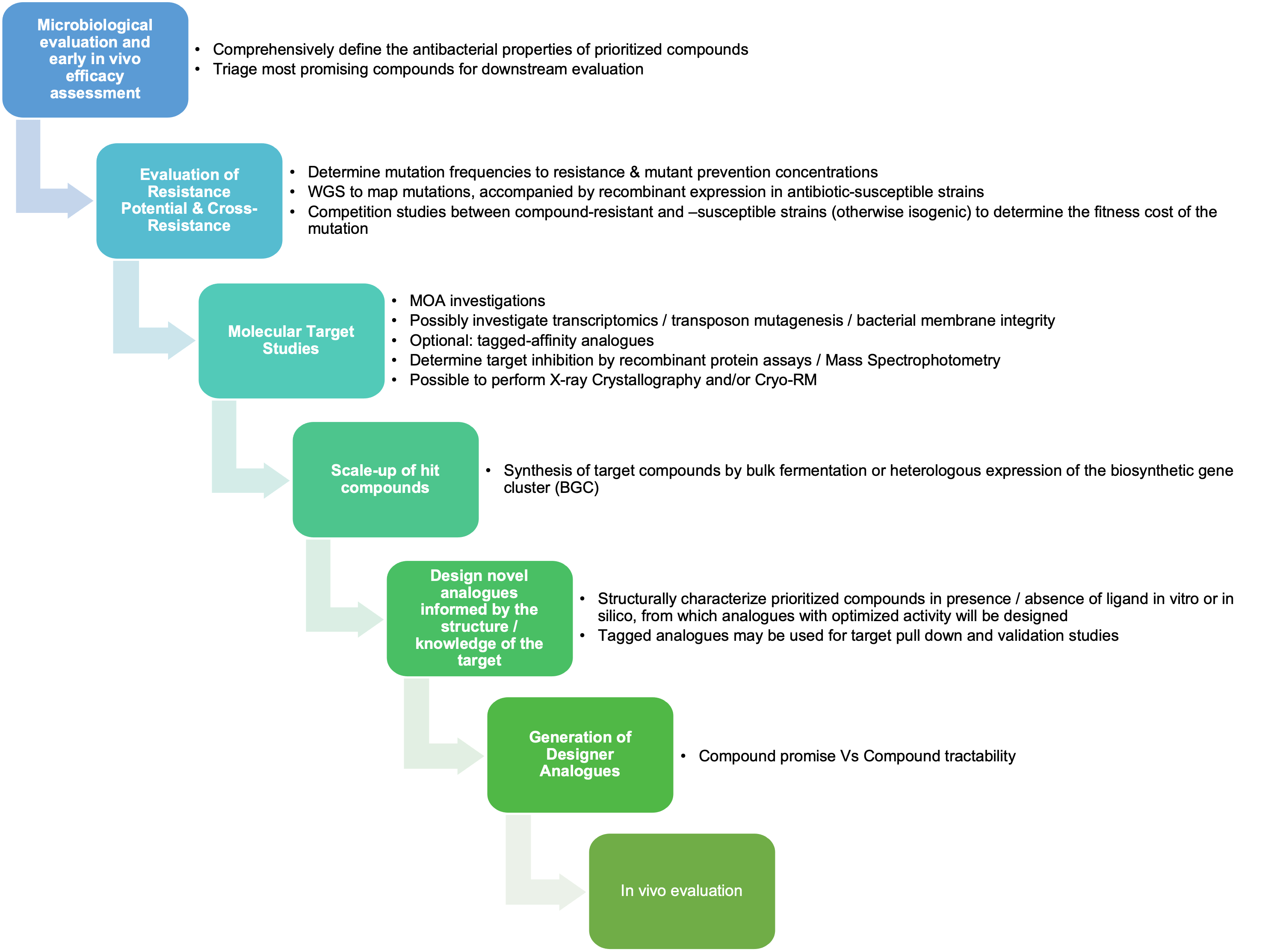
Hits identified and prioritised from Phase 1 (NP Discovery) (i.e. those compounds exhibiting promising antibacterial activity) will be progressed to Accelerator Phase 2 to establish a fully-functional ADD Hub. Promising hits will be characterized with respect to spectrum of activity, potency, prokaryotic selectivity, propensity for generating resistance, mode of action, in vivo efficacy.

Following the initial screening phase of Natural Product Discovery (Phase 1), the Project will enter Phase 2 - promising compounds identified in screening will be prioritized for further characterization and investigation.